3821161
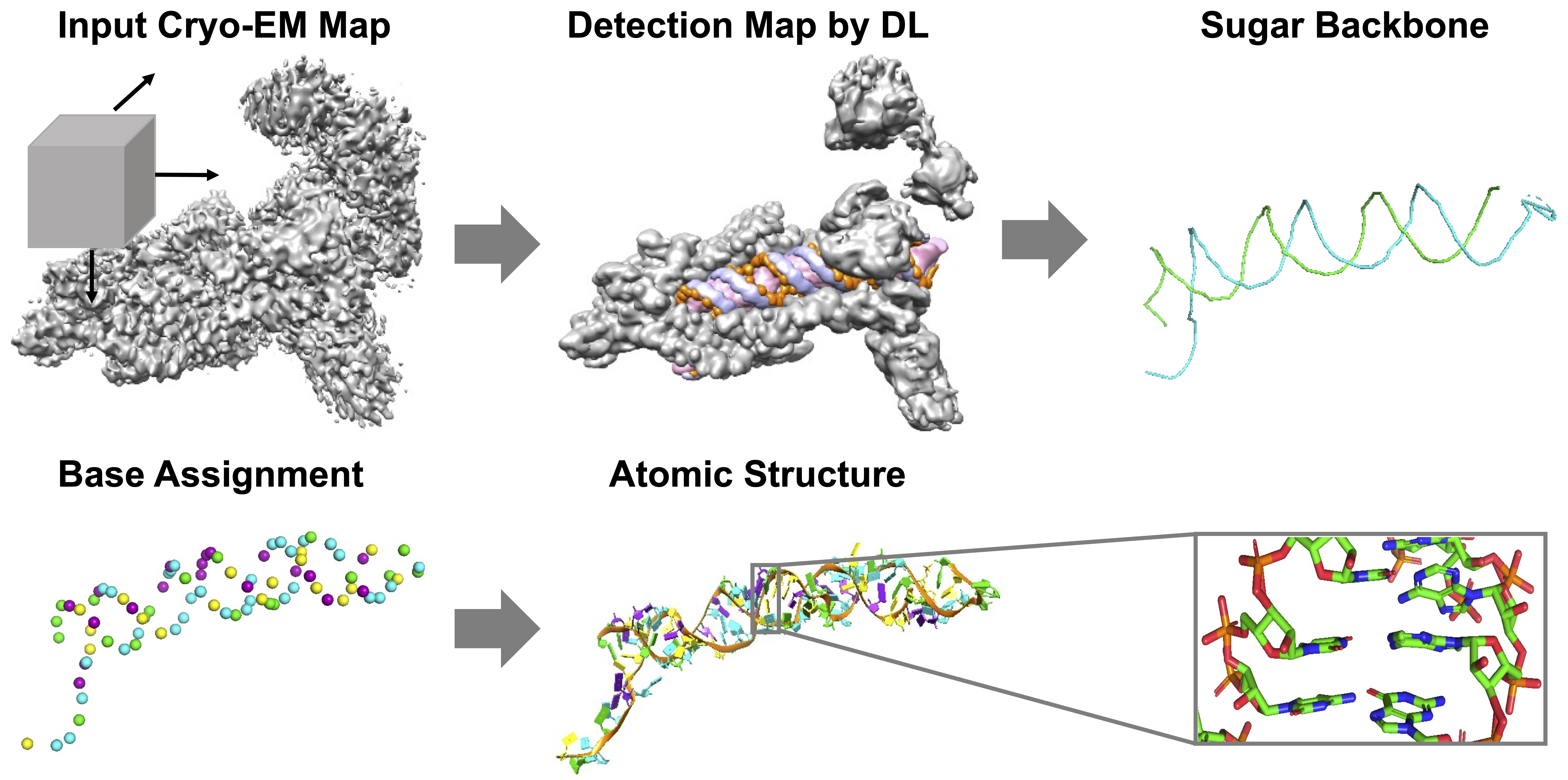
De novo structure modeling for nucleic acids in cryo-EM maps using deep learning | Poster Board #218
Date
March 28, 2023
Related Products
deepMainmast: De novo protein structure modeling for cryo-EM with deep learning and structure prediction | Poster Board #212
Structure modeling from density maps is a fundamental step for studying proteins and their complexes with cryogenic electron microscopy (cryo-EM)…
Protein complex structure modeling from Cryo-EM map using deep learning
An increasing numbers of protein complex structures have been determined with cryo-electron microscopy (cryo-EM)…
Computational & Theoretical Modeling of Biomolecular Dynamics in Crowded Environments: From Molecules to Cells: Machine Learning & Mathematical Models
DIVISION/COMMITTEE: [PHYS: Division of Physical Chemistry] [PHYS: Division of Physical Chemistry]
Understanding dynamic chromosome organization in living bacteria by single-molecule tracking and super-resolution microscopy
Nucleoid-associated proteins organize the bacterial chromosome and regulate gene expression; these proteins therefore contribute to the dynamic structure of the nucleoid across the bacterial lifespan…