Get the most out of ACS Spring 2025 by signing in with your ACS account.
Preferred
(EDT)
4191179 - Advancing proton-coupled electron transfer: Discovery of redox-active molecules with optimized Bdfes via constrained molecular generation
1:15 PM - 1:30 PM EDT
Thursday, March 27, 2025Room: Ballroom 6C (San Diego Convention Center)Parent Session
Generative Modeling for Chemistry, Biology, & Material Discovery:
Room: Ballroom 6C (San Diego Convention Center)
DIVISION/COMMITTEE: [CINF: Division of Chemical Information] [COMP: Division of Computers in Chemistry]
Credits
0.00 CE
Organizer, Presiders
- Connor Coley, Massachusetts Institute of Technology
- Chenru Duan
- Wenhao Gao, Massachusetts Institute of Technology
- Zhuoran Qiao, Chai Discovery
- Peichen Zhong, University of California Berkeley
Oral - In-person
COMP: Division of Computers in Chemistry
CINF: Division of Chemical Information
Overview
Proton-Coupled Electron Transfer (PCET) processes are essential for enhancing energy conversion, efficiency, and selectivity in chemical and biological systems, driving catalysis and sustainable energy solutions like fuel cells and redox flow batteries.
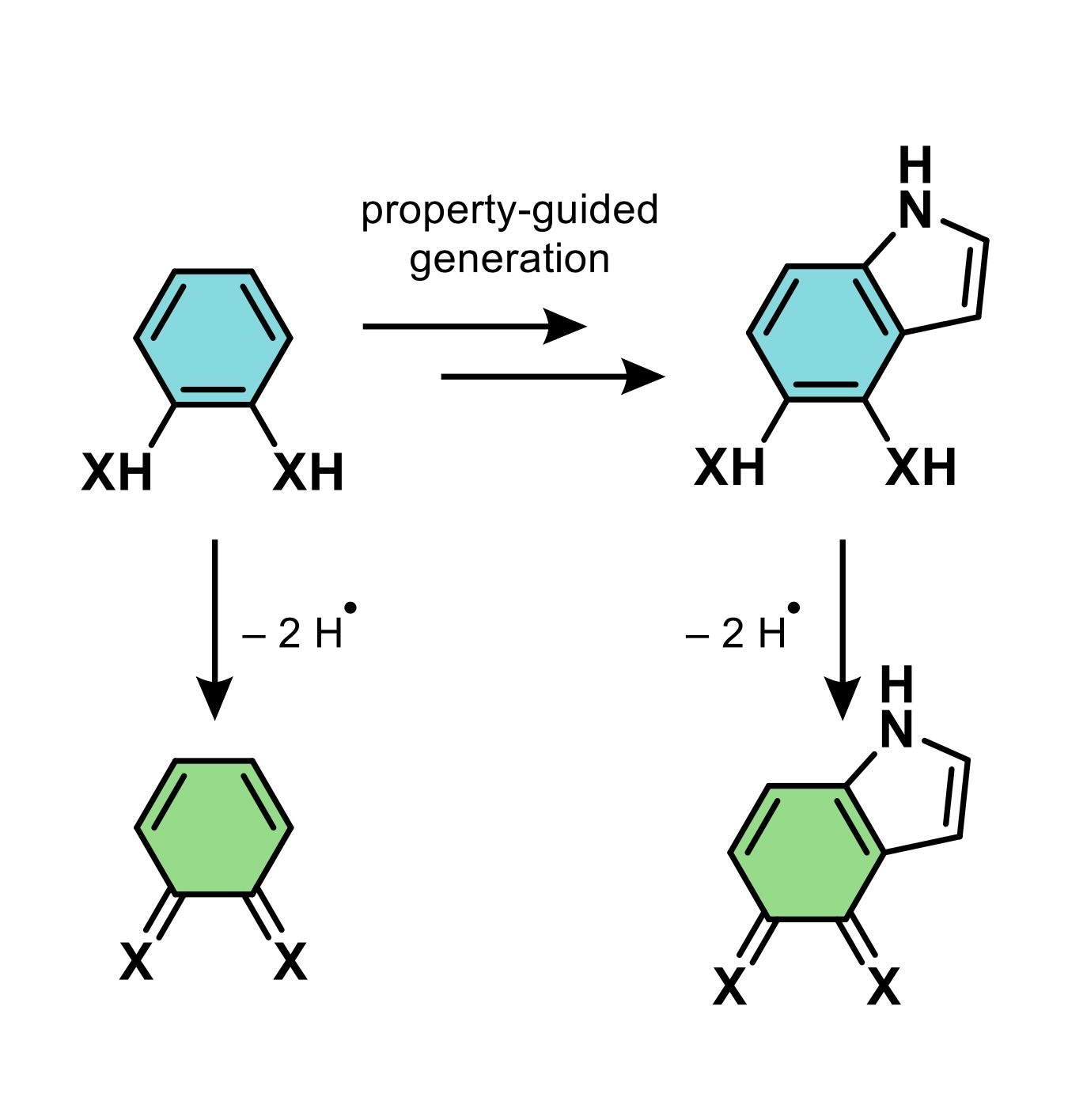
This work focuses on the thermodynamics, specifically bond dissociation free energy (BDFE), of redox-active molecules capable of PCET, based on the core scaffold shown in the attached scheme. While the thermochemical properties of hydroquinones and quinones (X = O) are well-
studied, analogous systems with X = NR’ or S remain underexplored. To address this, we developed a virtual screening workflow. AIMNET2, a neural potential from the Isayev group at CMU, accurately calculates BDFEs at DFT-level precision, enabling the creation of a virtual dataset of 30k diamines with BDFEs evaluated. A regression model trained on MolFormer encodings predicts AIMNET2 BDFEs with an R2 of 0.90, accelerating the discovery process.
For understudied core scaffolds such as 1,2-benzenedithiol and its derivatives, only around a thousand candidates have been identified out of the 110M molecules available in the PubChem repository. Here, we introduce the first family of graph neural networks capable of generating diverse yet scaffold-constrained molecules while minimizing the need for excessive post-hoc filtering. By integrating these models with genetic algorithms, we successfully expanded the candidate pool and identified molecules that fall within a desirable BDFE range.
This work has provided access to a broad spectrum of molecules, ranging from strong oxidizing agents (high BDFEs) to reducing agents (low BDFEs), as well as candidates for energy storage and conversion devices (intermediate BDFEs). Additionally, these organic molecules show promise as ligands for transition metal complexes, offering potential as catalysts for more challenging chemical transformations.
This work focuses on the thermodynamics, specifically bond dissociation free energy (BDFE), of redox-active molecules capable of PCET, based on the core scaffold shown in the attached scheme. While the thermochemical properties of hydroquinones and quinones (X = O) are well-
studied, analogous systems with X = NR’ or S remain underexplored. To address this, we developed a virtual screening workflow. AIMNET2, a neural potential from the Isayev group at CMU, accurately calculates BDFEs at DFT-level precision, enabling the creation of a virtual dataset of 30k diamines with BDFEs evaluated. A regression model trained on MolFormer encodings predicts AIMNET2 BDFEs with an R2 of 0.90, accelerating the discovery process.
For understudied core scaffolds such as 1,2-benzenedithiol and its derivatives, only around a thousand candidates have been identified out of the 110M molecules available in the PubChem repository. Here, we introduce the first family of graph neural networks capable of generating diverse yet scaffold-constrained molecules while minimizing the need for excessive post-hoc filtering. By integrating these models with genetic algorithms, we successfully expanded the candidate pool and identified molecules that fall within a desirable BDFE range.
This work has provided access to a broad spectrum of molecules, ranging from strong oxidizing agents (high BDFEs) to reducing agents (low BDFEs), as well as candidates for energy storage and conversion devices (intermediate BDFEs). Additionally, these organic molecules show promise as ligands for transition metal complexes, offering potential as catalysts for more challenging chemical transformations.
Presenter






